How Tob Read a Surface Data Plot
Lesson 7. Open, Plot and Explore Raster Data with Python
Important: We are in the process of updating our lessons to use rioxarray which wraps around rasterio to brand data processing easier. This lesson will be maintained for the future even so we are going to start education rasterio information processing using rioxarray.
Learning Objectives
- Open, plot, and explore raster data using Python.
- Handle no information values in raster information.
- Create plotting extents then you can plot raster and vector information together using matplotlib.
- Explore raster data using histograms and descriptive statistics.
Open Raster Data in Open up Source Python
Call up from the previous lesson that raster or "gridded" data are stored as a filigree of values which are rendered on a map as pixels. Each pixel value represents an surface area on the Earth's surface. A raster file is composed of regular grid of cells, all of which are the aforementioned size. Raster information can be used to shop many different types of scientific information including
- pinnacle data
- canopy superlative models
- surface temperature
- climate model data outputs
- landuse / landcover data
- and more.
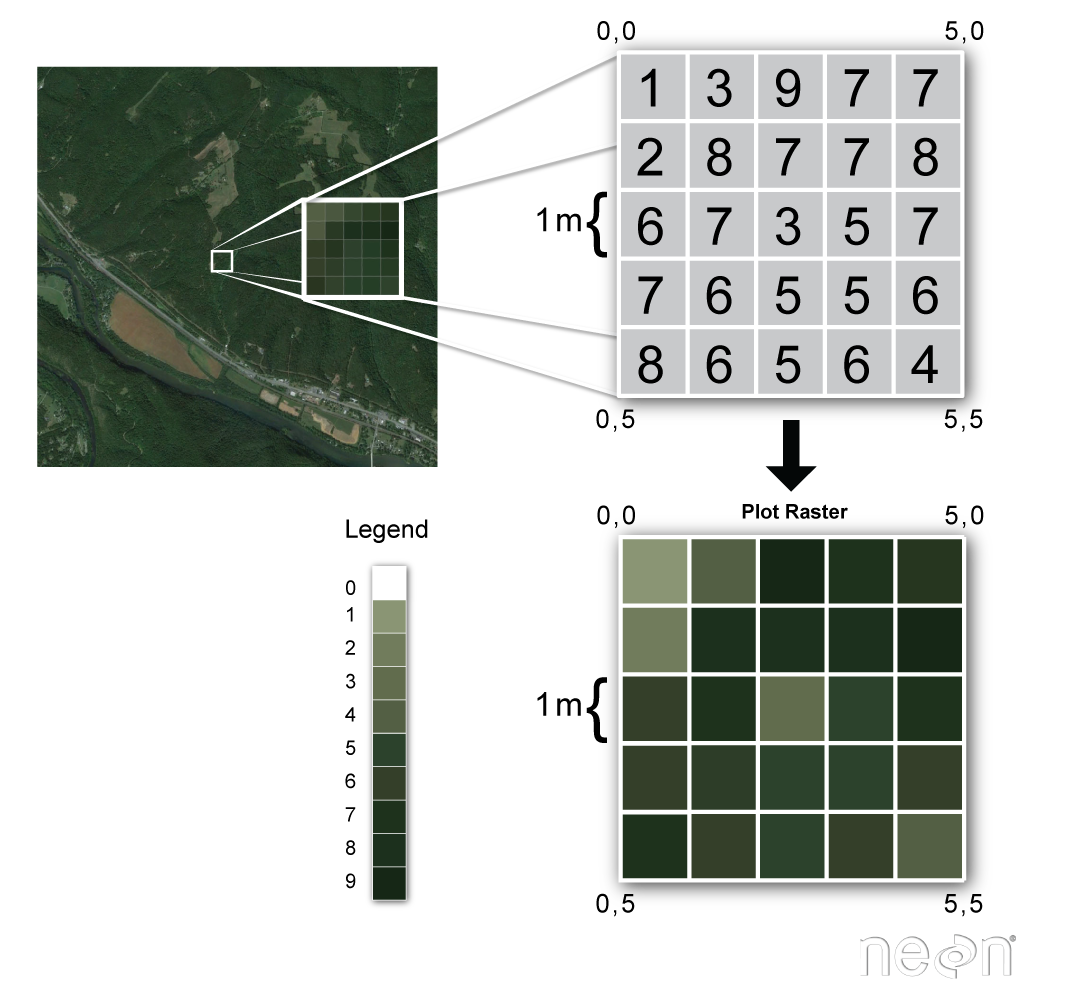
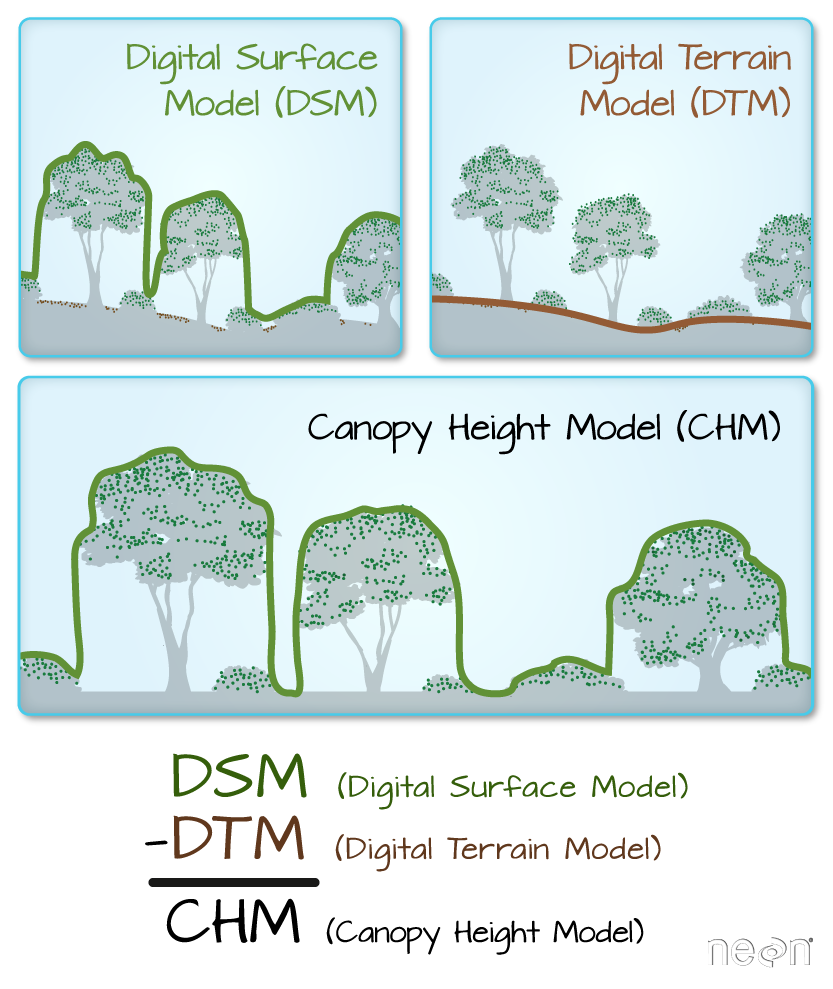
In this lesson you volition learn more well-nigh working with lidar derived raster information that represents both terrain / height data (elevation of the earth'south surface), and surface height (height at the tops of trees, buildings etc to a higher place the earth'southward surface). If you want to read more well-nigh how lidar data are used to derive raster based surface models, you can bank check out this chapter on lidar remote sensing data and the various raster data products derived from lidar data.
Information Tip: The information used in this lesson are NEON (National Ecological Observatory Network) data.
To begin load the packages that you need to process your raster information.
# Import necessary packages import bone import matplotlib.pyplot as plt import seaborn as sns # Use geopandas for vector data and rasterio for raster data import geopandas as gpd import rasterio equally rio # Plotting extent is used to plot raster & vector information together from rasterio.plot import plotting_extent import earthpy every bit et import earthpy.plot every bit ep # Prettier plotting with seaborn sns . set ( font_scale = ane.5 , style = "white" ) /opt/conda/lib/python3.8/site-packages/rasterio/plot.py:263: SyntaxWarning: "is" with a literal. Did you lot mean "=="? if len(arr.shape) is two: # Get data and set working directory et . data . get_data ( "colorado-flood" ) bone . chdir ( os . path . join ( et . io . HOME , 'earth-analytics' , 'information' )) Downloading from https://ndownloader.figshare.com/files/16371473 Extracted output to /root/world-analytics/data/colorado-alluvion/. Below, you define the path to a lidar derived digital elevation model (DEM) that was created using NEON (the National Ecological Observatory Network) data.
You then open the data using rio.open up("path-to-raster-here").
# Define relative path to file dem_pre_path = os . path . join ( "colorado-flood" , "spatial" , "boulder-leehill-rd" , "pre-flood" , "lidar" , "pre_DTM.tif" ) # Open the file using a context manager ("with rio.open up" statement) with rio . open ( dem_pre_path ) every bit dem_src : dtm_pre_arr = dem_src . read ( 1 ) When you open raster data using rasterio you are creating a numpy assortment. Numpy is an efficient way to work with and process raster format information. You can plot your data using earthpy plot_bands() which takes a numpy array as an input and generates a plot.
# Plot your data using earthpy ep . plot_bands ( dtm_pre_arr , championship = "Lidar Digital Elevation Model (DEM) \n Boulder Alluvion 2013" , cmap = "Greys" ) plt . show () 
The data above should represent terrain model data. However, the range of values is not what is expected. These data are for Bedrock, Colorado where the elevation may range from 1000-3000m.
There may exist some outlier values in the data that may demand to exist addressed. Below you check out the min and max values of the data.
impress ( "the minimum raster value is: " , dtm_pre_arr . min ()) print ( "the maximum raster value is: " , dtm_pre_arr . max ()) the minimum raster value is: -3.4028235e+38 the maximum raster value is: 2087.43 # A histogram tin can likewise be helpful to wait at the range of values in your data # What do yous notice most the histogram beneath? ep . hist ( dtm_pre_arr , figsize = ( ten , 6 )) plt . show () 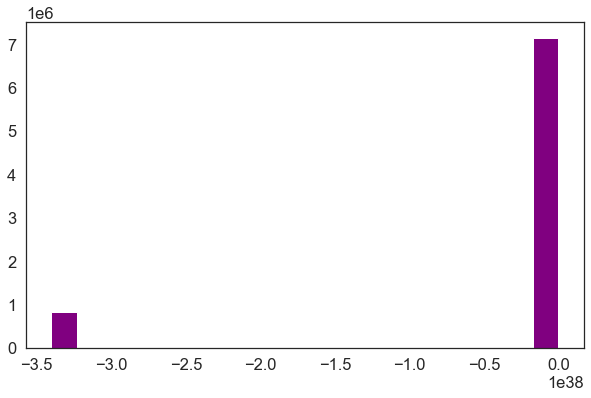
Raster Information Exploration - Min and Max Values
Looking at the minimum value of the data, in that location is one of ii things going on that need to be fixed
- there may be no data values in the data with a negative value that are skewing your plot colors
- in that location also could exist outlier data in your raster
You tin explore the kickoff choice - that there are no data values by reading in the data and masking no data values using rasterio. To do this, you will use the masked=Truthful parameter for the .read() office - similar this:
dem_src.read(one, masked=True)
# Read in your data and mask the no data values with rio . open ( dem_pre_path ) as dem_src : # Masked=True will mask all no data values dtm_pre_arr = dem_src . read ( 1 , masked = True ) Observe that now the minimum value looks more than like an elevation value (which should almost often not be negative).
print ( "the minimum raster value is: " , dtm_pre_arr . min ()) print ( "the maximum raster value is: " , dtm_pre_arr . max ()) the minimum raster value is: 1676.21 the maximum raster value is: 2087.43 # A histogram tin also be helpful to expect at the range of values in your data ep . hist ( dtm_pre_arr , figsize = ( 10 , 6 ), title = "Histogram of the Information with No Data Values Removed" ) plt . testify () 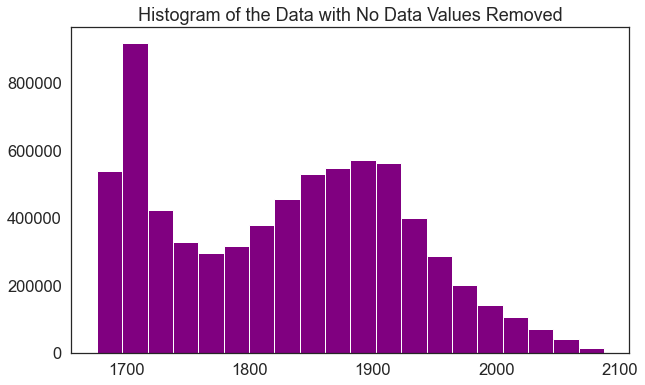
Plot your data again to see how it looks.
# Plot data using earthpy ep . plot_bands ( dtm_pre_arr , title = "Lidar Digital Elevation Model (DEM) \n Boulder Overflowing 2013" , cmap = "Greys" ) plt . show () 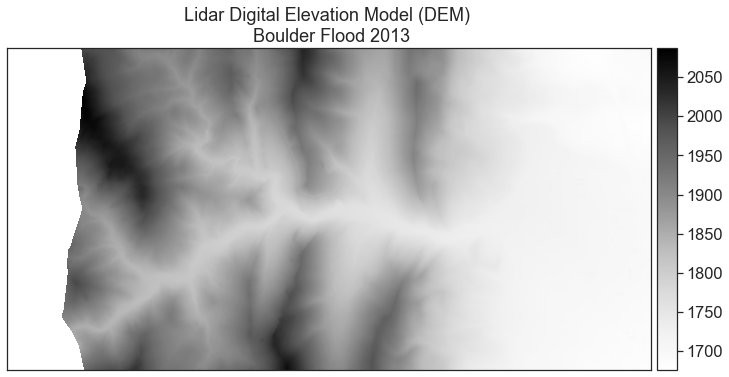
Rasterio Reads Files into Python equally Numpy Arrays
When you call src.read() above, rasterio is reading in the data as a numpy array. A numpy assortment is a matrix of values. Numpy arrays are an efficient construction for working with large and potentially multi-dimensional (layered) matrices.
The numpy array beneath is type numpy.ma.core.MaskedArray. It is a masked array because you chose to mask the no data values in your data. Masking ensures that when you lot plot and perform other math operations on your information, those no information values are not included in the operations.
Data Tip: If yous want to learn more nigh Numpy arrays, check out the intro to earth datascience textbook chapter on Numpy arrays.
with rio . open ( dem_pre_path ) as dem_src : lidar_dem_im = dem_src . read ( 1 , masked = Truthful ) print ( "Numpy Array Shape:" , lidar_dem_im . shape ) impress ( "Object type:" , type ( lidar_dem_im )) Numpy Array Shape: (2000, 4000) Object blazon: <class 'numpy.ma.core.MaskedArray'> A numpy array does not past default store spatial information. However, your raster information is spatial - it represents a location on the earth'south surface.
You tin acccess the spatial metadata within the context manager using dem_src.profile. Detect that the .contour object contains information including the no data values for your data, the shape, the file type and fifty-fifty the coordinate reference system. You will learn more about raster metadata in the raster metadata lesson in this chapter.
with rio . open ( dem_pre_path ) as dem_src : lidar_dem_im = dem_src . read ( one , masked = True ) # Create an object called lidar_dem_meta that contains the spatial metadata lidar_dem_meta = dem_src . profile lidar_dem_meta {'commuter': 'GTiff', 'dtype': 'float32', 'nodata': -3.4028234663852886e+38, 'width': 4000, 'meridian': 2000, 'count': one, 'crs': CRS.from_epsg(32613), 'transform': Affine(ane.0, 0.0, 472000.0, 0.0, -i.0, 4436000.0), 'blockxsize': 128, 'blockysize': 128, 'tiled': True, 'compress': 'lzw', 'interleave': 'band'} Context Managers to Open up and Close File Connections
The steps in a higher place correspond the steps yous need to open and plot a raster dataset using rasterio in python. The with rio.open up() statement creates what is known equally a context manager. A context manager allows yous to open the information and work with information technology. Within the context manager, Python makes a temporary connection to the file that you lot are trying to open up.
In the example above this was a file chosen pre_DTM.tif.
To break this code down, the context director has a few parts. First, information technology has a with statement. The with statement creates a connection to the file that you want to open up. The default connexion blazon is read only. This means that you lot tin Non modify that file by default. Not being able to change the original data is a good affair because information technology prevents yous from making unintended changes to your original information.
with rio.open(`file-path-here`) equally file_src: dtm_pre_arr = dem_src.read(1, masked=True) Discover that the beginning line of the context manager is not indented. It contains two parts
-
rio.open(): This is the code that will open a connection to your .tif file using a path you provide. -
file_src: this is a rasterio reader object that you tin can use to read in the actual data. You can too employ this object to access the metadata for the raster file.
The second line of your with argument
dtm_pre_arr = dem_src.read(1, masked=True)
is indented. Any code that is indented directly below the with argument volition become a part of the context manager. This code has direct admission to the file_src object which is y'all recall above is the rasterio reader object.
Opening and closing files using rasterio and context managers is efficient every bit information technology establishes a connection to the raster file rather than directly reading it into retentivity.
In one case you lot are washed opening and reading in the data, the context manager closes that connection to the file. This efficiently ensures that the file won't be modified later on in your code.
Data Tip: You can open and shut files without a context manager using the syntax below. This arroyo nevertheless is generally non recommended.
lidar_dem = rio.open(dem_pre_path) lidar_dem.close() You lot can go a meliorate understanding of how the rasterio context director works by taking a wait at what it is doing line by line. Start by looking at the dem_pre_path object. Notice that this object is a path to the file pre_DEM.tif. The context managing director needs to know where the file is that you lot desire to open up with Rasterio.
# Look at the path to your dem_pre file dem_pre_path 'colorado-flood/spatial/boulder-leehill-rd/pre-flood/lidar/pre_DTM.tif' Now utilize the dem_pre_path in the context director to open and close your connexion to the file. Notice that if you print the "src" object within the context manager (detect that the print statement is indented which is how you know that you are within the context director), the returl is an
open DatasetReader
The name of the reader is the path to your file. This ways there is an open and agile connexion to the file.
# Opening the file with the dem_pre_path # Notice hither the src object is printed and returns an "open" DatasetReader object with rio . open ( dem_pre_path ) as src : print ( src ) <open up DatasetReader name='colorado-flood/spatial/boulder-leehill-rd/pre-alluvion/lidar/pre_DTM.tif' mode='r'> If you print that aforementioned src object outside of the context managing director, notice that it is now a closed datasetReader object. Information technology is airtight because it is being chosen outside of the context manager. Once the connection is airtight, y'all tin no longer access the data. This is a good thing as information technology protects you lot from inadvertently modifying the file itself!
# Note that the src object is now airtight because it'south not within the indented # part of the context manager above print ( src ) <closed DatasetReader name='colorado-flood/spatial/boulder-leehill-rd/pre-overflowing/lidar/pre_DTM.tif' style='r'> Now look at what .read() does. Below you use the context manager to both open the file and read it. See that the read() method, returns a numpy array that contains the raster cell values in your file.
# Open up the file using a context managing director and get the values as a numpy array with .read() with rio . open ( dem_pre_path ) as dem_src : dtm_pre_arr = dem_src . read ( ane ) dtm_pre_arr array([[-3.4028235e+38, -three.4028235e+38, -3.4028235e+38, ..., ane.6956300e+03, i.6954199e+03, 1.6954299e+03], [-3.4028235e+38, -3.4028235e+38, -three.4028235e+38, ..., i.6956000e+03, 1.6955399e+03, ane.6953600e+03], [-3.4028235e+38, -iii.4028235e+38, -3.4028235e+38, ..., ane.6953800e+03, 1.6954399e+03, i.6953700e+03], ..., [-three.4028235e+38, -3.4028235e+38, -three.4028235e+38, ..., 1.6814500e+03, one.6813900e+03, 1.6812500e+03], [-3.4028235e+38, -3.4028235e+38, -three.4028235e+38, ..., 1.6817200e+03, 1.6815699e+03, 1.6815599e+03], [-3.4028235e+38, -3.4028235e+38, -three.4028235e+38, ..., ane.6818900e+03, 1.6818099e+03, 1.6817400e+03]], dtype=float32) Because you lot created an object within the context managing director that contains those raster values equally a numpy array, you lot can now admission the data values without needing to have an open connection to your file. This ensures once more that you are not modifying your original file and that all connections to it are closed. You are now free to play with the numpy array and procedure your data!
# View numpy assortment of your information dtm_pre_arr array([[-3.4028235e+38, -3.4028235e+38, -3.4028235e+38, ..., 1.6956300e+03, i.6954199e+03, ane.6954299e+03], [-3.4028235e+38, -3.4028235e+38, -three.4028235e+38, ..., 1.6956000e+03, ane.6955399e+03, 1.6953600e+03], [-3.4028235e+38, -3.4028235e+38, -3.4028235e+38, ..., i.6953800e+03, 1.6954399e+03, 1.6953700e+03], ..., [-3.4028235e+38, -iii.4028235e+38, -3.4028235e+38, ..., 1.6814500e+03, 1.6813900e+03, 1.6812500e+03], [-3.4028235e+38, -3.4028235e+38, -iii.4028235e+38, ..., i.6817200e+03, 1.6815699e+03, one.6815599e+03], [-three.4028235e+38, -iii.4028235e+38, -iii.4028235e+38, ..., 1.6818900e+03, 1.6818099e+03, one.6817400e+03]], dtype=float32) You tin can use the .contour attribute to create an object with metadata on your raster image. The metadata object below contains information similar the coordinate reference arrangement and size of the raster image.
with rio . open up ( dem_pre_path ) as dem_src : # Create an object chosen lidar_dem_meta that contains the spatial metadata lidar_dem_meta = dem_src . profile lidar_dem_meta {'driver': 'GTiff', 'dtype': 'float32', 'nodata': -3.4028234663852886e+38, 'width': 4000, 'height': 2000, 'count': 1, 'crs': CRS.from_epsg(32613), 'transform': Affine(one.0, 0.0, 472000.0, 0.0, -ane.0, 4436000.0), 'blockxsize': 128, 'blockysize': 128, 'tiled': True, 'compress': 'lzw', 'interleave': 'band'} Finally, accept a look at what the plotting_extent() role does. Note below that when you lot run the plotting_extent() function on your dem_pre raster image, you lot get the extent of the image as an output.
with rio . open ( dem_pre_path ) equally dem_src : # Create an object chosen lidar_dem_plot_ext that contains the spatial metadata lidar_dem_plot_ext = plotting_extent ( dem_src ) # This plotting extent object will exist used below to ensure your data overlay correctly lidar_dem_plot_ext (472000.0, 476000.0, 4434000.0, 4436000.0) Plot Raster and Vector Information Together: Plot Extents
Numpy arrays are an efficient mode to store and process data. Yet, by default they do not contain spatial information. To plot raster and vector information together on a map, you will need to create an extent object that defines the spatial extent of your raster layer. This volition then allow you to plot a raster and vector data together to create a map.
Beneath you open a single shapefile that contains a boundary layer that y'all can overlay on elevation of your raster dataset.
# Open site boundary vector layer site_bound_path = bone . path . join ( "colorado-flood" , "spatial" , "boulder-leehill-rd" , "prune-extent.shp" ) site_bound_shp = gpd . read_file ( site_bound_path ) # Plot the vector data site_bound_shp . plot ( color = 'teal' , edgecolor = 'black' ) plt . show () 
Y'all can try to plot the two datasets together merely you will see below that the output plot does not look correct. This is because the raster layer has no spatial information associated with it.
fig , ax = plt . subplots ( figsize = ( iv , 10 )) ep . plot_bands ( dtm_pre_arr , ax = ax ) site_bound_shp . plot ( color = 'teal' , edgecolor = 'black' , ax = ax ) plt . bear witness () 
with rio . open ( dem_pre_path ) equally dem_src : lidar_dem_im = dem_src . read ( i , masked = Truthful ) # Create an object called lidar_dem_plot_ext that contains the spatial metadata lidar_dem_plot_ext = plotting_extent ( dem_src ) # This plotting extent object volition be used beneath to ensure your data overlay correctly lidar_dem_plot_ext (472000.0, 476000.0, 4434000.0, 4436000.0) Next endeavour to plot. This time however, employ the extent= parameter to specify the plotting extent within ep.plot_bands()
fig , ax = plt . subplots () ep . plot_bands ( dtm_pre_arr , ax = ax , extent = lidar_dem_plot_ext ) site_bound_shp . plot ( colour = 'None' , edgecolor = 'teal' , linewidth = 2 , ax = ax ) # Plow off the outline or axis border on your plot ax . centrality ( 'off' ) plt . prove () 
Information Tip: Customizing Raster Plot Color Ramps To change the color of a raster plot yous set the colormap. Matplotlib has a list of pre-adamant color ramps that yous can chose from. You can reverse a color ramp by calculation _r at the end of the color ramp's name, for instance cmap = 'viridis' vs cmap = 'viridis_r'.
You lot at present have the basic skills needed to open and plot raster data. Complete the challenges below to exam your skills.
chabrillanannothing.blogspot.com
Source: https://www.earthdatascience.org/courses/use-data-open-source-python/intro-raster-data-python/fundamentals-raster-data/open-lidar-raster-python/
0 Response to "How Tob Read a Surface Data Plot"
Post a Comment